Biological role of Non Canonical D-Amino Acids (NCDAAs)
Production of NCDAAs in Vibrio cholerae is performed by BsrV, a new type of multi-specific racemase. Given the important adaptive properties provided by NCDAAs to cell wall and biofilm stability, investigation of this type of racemases in detail is critical for both understanding NCDAAs controlled processes as for the biotechnological and biomedical exploitation of these biocatalysts.
In addition, BsrV expression is fully dependent on RpoS, suggesting that NCDAAs are released in all environmental situations that require this stress sigma factor. A major goal of our lab is to identify the biological benefit that production of NCDAA gives to Bsr-encoding bacteria at all levels.
Characterization of the peptidoglycan-remodeling pathways by NCDAAs
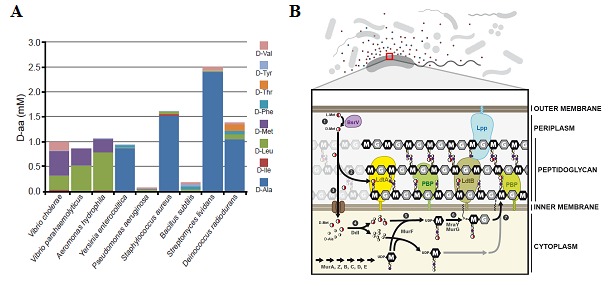
PG regulation by NCDAAs occurs mainly through the generation of modified muropeptides, which, acting as non-preferred substrates, might slow down the activity of the PBPs and, thus, PG biosynthesis rate. In a previous work, we unveiled two enzymatic pathways for NCDAA incorporation: periplasmic LD-transpeptidases (Ldt) and/or cytoplasmic biosynthetic ligases (Ddl and MurF).
We want to characterize these activities and the consequences of such editing on the enzymes that synthesize and remodel the murein sacculus. Elucidating the mechanisms by which NCDAAs govern cell wall remodelling will undoubtedly reveal new paradigms for understanding how extra-cytoplasmic processes are regulated as well as lead to development of novel therapeutics.
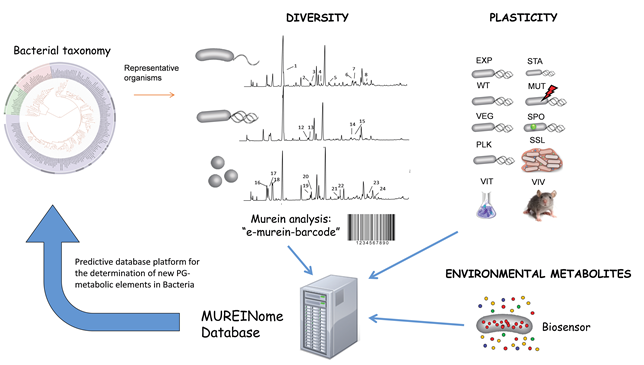
Beyond NCDAAs: Analysis of diversity and plasticity of the bacterial PG
We have launched an integrative research program to uncover and exploit the unnoticed variability of the bacterial cell wall. This investigation is imperative for a realistic understanding of cell wall biology in nature, in particular its role in environmental adaptation and signalling.
Gathered data will be the basis of the first comprehensive bacterial cell wall database and ad hoc software: The MUREINome. Such a database will be highly relevant to properly understand bacterial relations with neighbouring organisms at all levels, and their adaptation to environmental challenges.
Moreover, this research might lead to the discovery of new metabolic and regulatory pathways with great potential in the development of new species-specific antimicrobials therapies. Therefore, this project promotes collaborative networks amongst fairly unconnected disciplines in life sciences, both in academia and in the public health field.
META-MUREINOME: A snap shot of bacterial cell wall environmental diversity

Despite bacteria being the most versatile organisms on Earth, most species cannot be investigated in the laboratory since their growth is strictly dependent on the activity of neighboring organisms that share the same niche. To overcome this drawback, different –omics have introduced global analysis (META-analysis) of genomes, proteins, RNA and metabolites to perceive the presence of microbes and the dynamics of their processes right in the natural environment.
Similarly, cell wall research can only be interpreted correctly if laboratory bench work is reconciled with environmental analysis. We propose to decipher the META-MUREINOME of specific, well defined environments as the starting point of what could develop into a new scientific field.
Temperature and metabolic dependent regulation of biofilm in Vibrio cholerae
A common strategy for bacterial growth and survival is the formation of biofilms – bacterial communities found in surfaces and interfaces held together by an extracellular matrix. The formation and subsequent dispersal of the biofilm is of major importance for colonization of a host and the generation of outbreaks.
Recent evidence shows that biofilm development can be influenced by signalling molecules, i.e. secondary metabolites that bacteria produce and secrete to the environment and that play an important role both in intra- and inter-species communication. This is of great importance in the environment, where bacteria must develop strategies to compete with other bacteria inhabiting the same niche, adapt to ever-changing conditions and survive.
Our group is interested in deciphering molecular mechanisms behind environmental stimuli that govern biofilm development of V. cholerae which could be of importance (i) upon infection of a host and subsequent transmission, and (ii) for interspecies communication with other microorganisms inhabiting the same niche in their natural environment.
Vibrio cholerae (left) and Pseudomonas aeruginosa (right) colonies.
Genetic determination of Vibrio cholerae cell wall homeostasis and morphogenesis
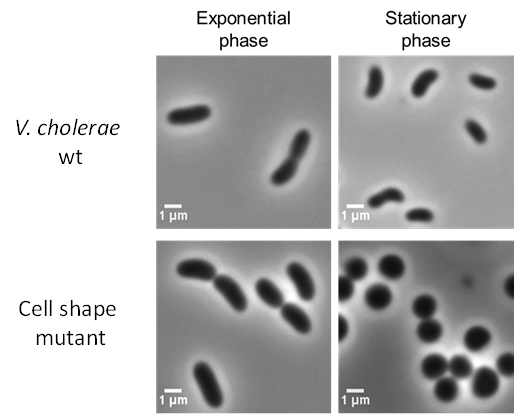
There are many fundamental cellular processes in bacteria (i.e cell wall biosynthesis) are often regulated by multiple genetic elements. This functional redundancy protects cell’s viability from genetic or chemical inactivation. V. cholerae cells have a characteristic curved rod-shape morphology, similar to those present in other gastrointestinal pathogens like Helicobacter pylori and Campylobacter jejuni.
Recent studies revealed novel factors (e.g. peptidoglycan hydrolytic enzymes) regulating helical shape in these bacteria and have provided unprecedented insights that link morphogenesis and bacterial adaptation to infection. Hence it is of clinical importance to uncover new factors regulating bacterial shape and polarity.
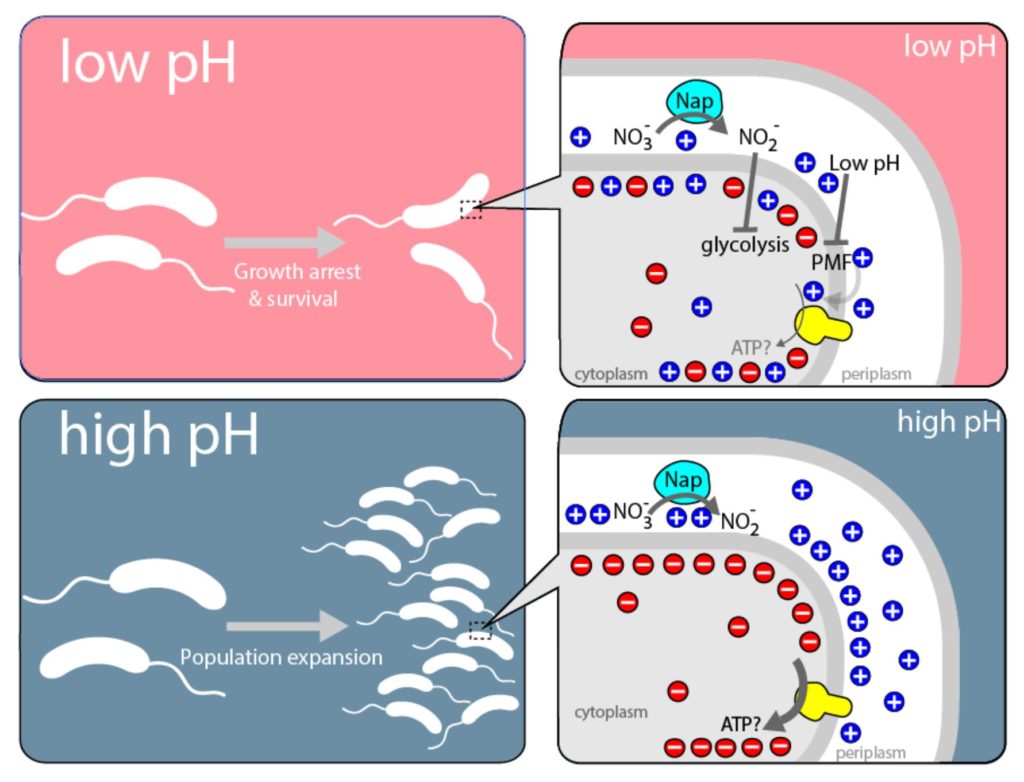
Metabolic switches controlling bacterial populations
The human pathogen Vibrio cholerae has puzzled the scientists since its discovery 150 years ago. Experts who studied the bacterium couldn’t understand that the bacterium was not able to grow under anaerobic conditions despite it was equipped with an active metabolic machinery which should enable the organism to respire nitrate instead of oxygen, conditions that typically exist in the gastrointestinal tract. The common opinion was that the bacteria accumulates the intermediate product nitrite which inhibits further growth.
We are studying this bacteria under low-oxygen and different pH-conditions. We have discovered an elegant pH-dependent metabolic mechanism which permits the pathogen to switch to a resting mode with preserved viability, providing competitive advantage against commensal bacteria to better colonize and infect the intestine.